Content Status
Type
Linked Node
A batch of samples is defined as a group of samples that are processed, amplified and hybridized:
- At the same time
- Under the same conditions
- Using the same PCR master mix, thermal cycler and hybridization solutions
- On the same hybridization platform.
QC during the DNA extraction procedure
- Use two sets of 1.5 ml tubes are foreach specimen.
- Mark and label the tubes according to the organization of the batch on the worksheet.
- The first tube in every batch should be labelled with a coloured sticker.
- Check the functionality of the instruments prior to processing.
- Use calibrated pipettes.
QC during the preparation of the master mix
Use homogenous reagents and thaw them properly to ensure even distribution of contents.
- Use calibrated pipettes.
- Set functional refrigerator/freezer at -20ºC.
- Verify addition of DNA polymerase visually.
- Document lot numbers and expiration dates of reagents.
- Test quality control specimens with a new lot of DNA polymerase.
QC during DNA template addition
- Select thermal cycler program based on the type of specimen to be amplified.
- Check that the caps of PCR tubes do not contain any liquid when loaded into the thermal cycler (to prevent dilution of the reaction mixture).
- Check the PCR reaction tubes for any bubbles (prevent uneven temperature distribution).
QC during the hybridization and detection procedure
- Maintain consistency in the numbering process. For example, strip marked number 1 must correspond with the first specimen extracted, amplified and added to denaturing buffer in hybridization well.
- Use dedicated pair of forceps to handle unhybridized strips.
- Avoid strips sticking together into the same well.
- If an automated hybridiser is used for less than 48 specimens, the rest of the wells should be filled with distilled water to ensure an even temperature across the heating platform.
- TwinCubator (12 strip capacity) should be used with one or two hybridization runs depending on the number of specimens.
- Use a clean tray in the TwinCubator.
Use of positive and negative controls
To demonstrate competency for the line probe assay (LPA), positive and negative quality control (QC) samples must be performed on a routine basis, for each batch of specimens as shown in Figure 1. The overview of the use of positive and negative quality control is shown in Figure 2.
Each batch of specimens tested with the LPA must have an “extraction positive control” (ATCC strain H37Rv), an “extraction negative control”, and a “PCR master mix negative control” (Figure 1).

Figure 1: The H37Rv extraction positive control shown in strip number 9 (circled in blue) and the extraction negative control is shown in strip number 10 (circled in red); Source: LPA Laboratory Manual, FIND
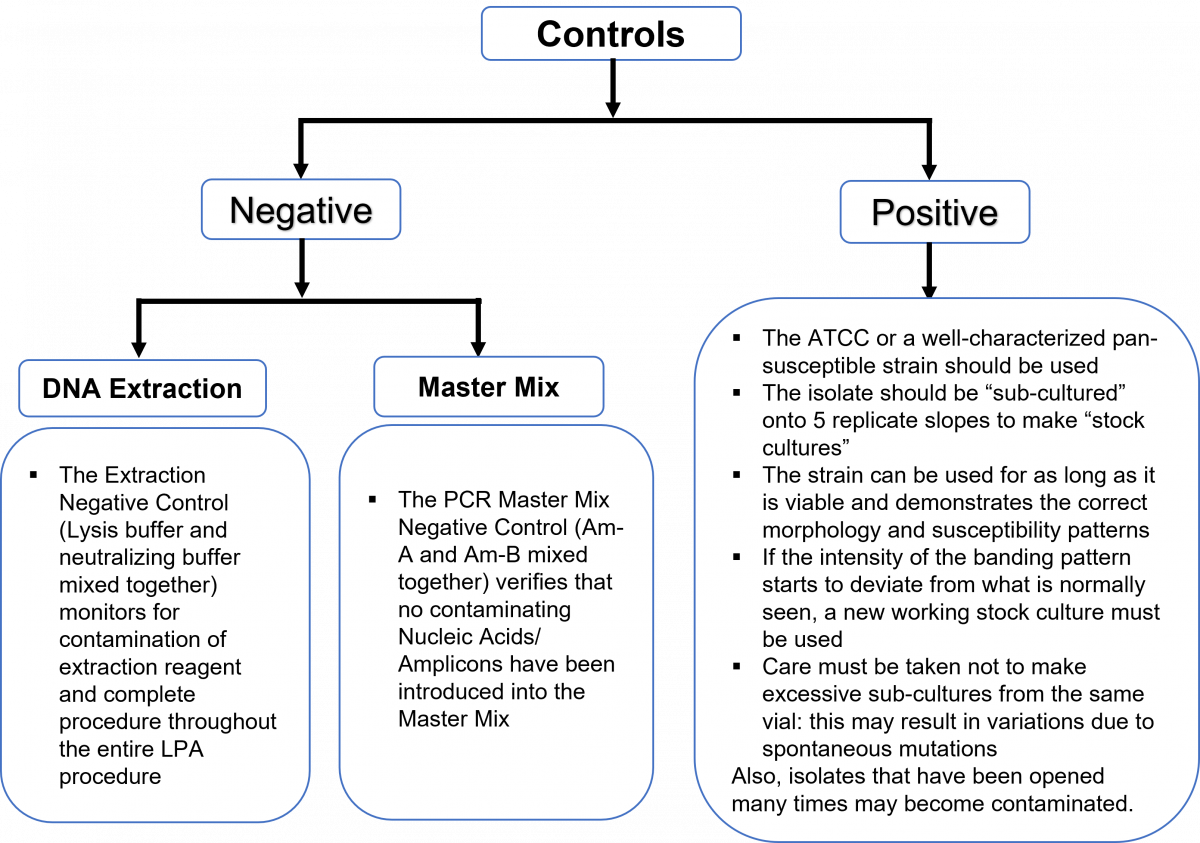
Figure 2: Overview of positive and negative controls
Amplification Control (AC) on the LPA strip functions as both:
The internal “PCR Positive Control”
Amplification control (AC) band indicates that the DNA extraction and PCR procedures were carried out successfully (Figure 3).
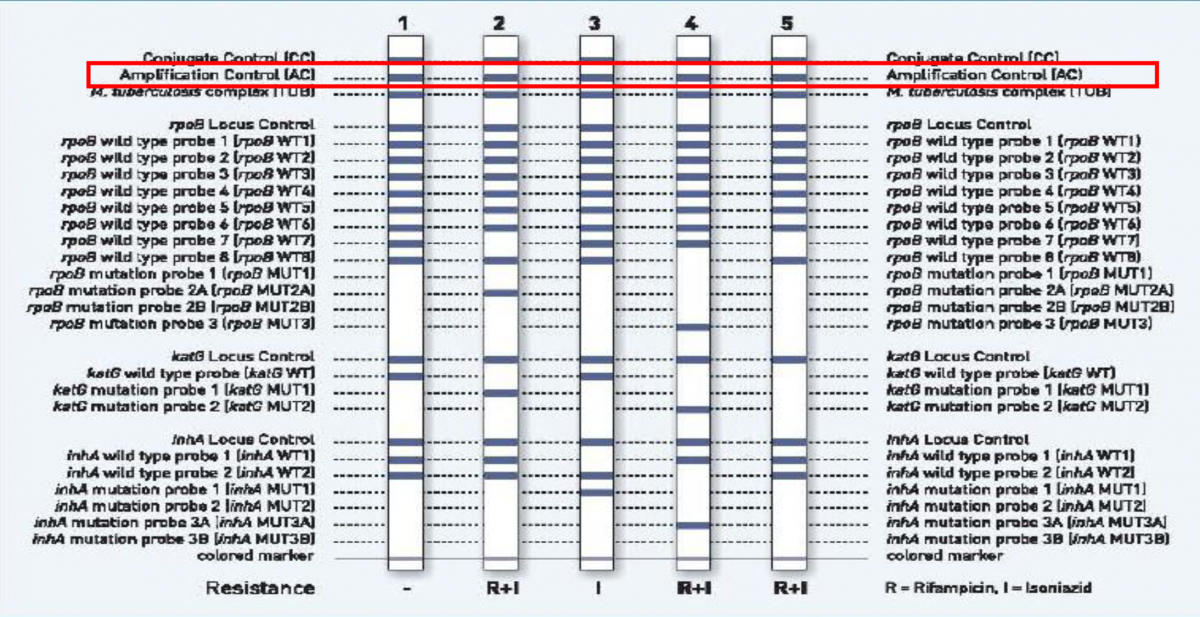
Figure 3: Amplification Control (highlighted in red) as seen on the Genotype MTBDRplus test strip; Source: LPA Laboratory Manual, FIND.
The “Inhibition Positive Control”
- The AC band will not appear if there are PCR inhibitors in the extracted material.
- The strips for the two negative controls must be positive only at the Conjugate Control (CC) and AC bands.
Resources
Kindly provide your valuable feedback on the page to the link provided HERE
Content Creator
Reviewer
- Log in to post comments